report
Abstract
The transition from the Digital Botanical Garden Initiative (DBGI) pilot project to the main Earth Metabolome Initiative (EMI) is a very important step towards adapting our collection and extraction methods to a wider spectrum of organisms or groups of specimens. This transitional project, undertaken as a Bachelor's thesis at the University of Fribourg, involves a comparative study of the metabolomic content of crops, weeds and invertebrates present in two different fields, one cultivated according to a biodynamic farming system and the other using conventional agriculture and only mineral fertilizers. The main objective of this study is not the metabolomic results themselves, although they are very interesting, but rather the adaptation of methods initially designed for data collection in a botanical garden to less controlled environments, with the aim of eventually setting up collection and extraction methods feasible in wild environments. This methodological adaptation is crucial to the successful scaling-up of the EMI, ensuring that robust and reliable data can be collected in diverse and variable natural environments, thereby facilitating the achievement of the initiative's overall objectives.
Background
Earth metabolome initiative
The Earth Metabolome Initivative (EMI) is a large scale open science project whose aim is to profile the metabolomic content of all organisms currently known on the planet Earth. This initiative have three main objectives : The first one is to protect biodiversity by creating molecular and metabolomic arguments, the second is to understand the chemical functioning of the biosphere and the last one is to benefit human society, notably through drug discovery or food sustainability. Understanding the chemical functioning of the biosphere is fundamental to developing effective conservation strategies. This understanding enables us to better protect biological diversity and guarantee the sustainability of the ecosystem services on which the survival of all species, including our own, and our well-being depend. Establishing a global molecular map of chemical content could help us to deep into new concepts such as "chemical endemism" or create some argument based on the chemodiversity level of certain zones. Ultimately, this will create a library accessible to all, enabling targeted research and protection of certain organisms and metabolites. These data will enable us to observe the chemodiversity of specific locations and thus add chemical arguments to environmental protection.
https://doi.org/10.5281/zenodo.10663625 https://doi.org/10.5281/zenodo.7486984
Digital botanical gardens initiative
Background
The digital botanical garden Initiative (DBGI) is an EMI pilot project whose aim is to extract, catalogue, analyze and create a digital library of metabolomic data from plants found in botanical gardens. This enables the methods to be tested and the bioinformatics resources needed for data processing to be created.
Why choose the botanical gardens for this pilot project? Botanical gardens, although human-made, are hotspots of biodiversity and the ideal place to test collection, extraction and analysis methods. They display a considerable number of plants in a small, accessible area near to university and laboratories supplies. Botanical gardens provide an initial approach and the methods needed to eventually collect different organisms from wild ecosystems (see Earth Metabolome Initiative). Their biodiversity also makes it possible to set up a library of chemical extracts, enabling the development of the IT tools needed to analyze these data.
Fribourg is privileged to have a university botanical garden where over 5,000 species are cultivated and conserved. This represents one of the largest systemic collections in Switzerland. The botanical garden and its research group play an active role in the conservation of rare and endangered species at cantonal and national level. At this point, around 2400 plants have already been collected, 1500 have been extracted and 1300 have been profiled with liquid chromatography mass spectrometry.
Tropical greenhouse
The tropical greenhouse (number 1 on the map) was sampled during my bachelor work. This greenhouse features decorative green plants, climbers, epiphytes and aquatic plants, all of which come from the rainforest. In all, some 280 plants are present with no specific taxonomic or provenance links.
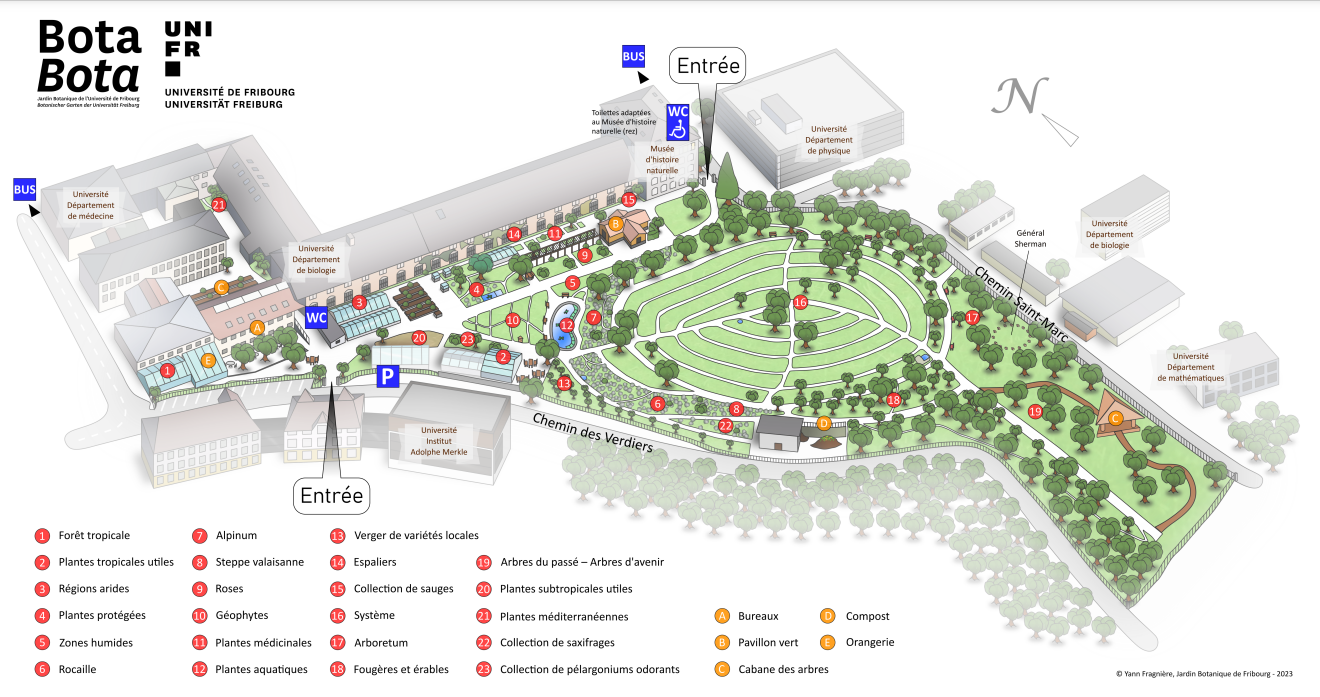 Map of Fribourg botanical garden. The tropical green house is The tropical greenhouse is located at point 1
Map of Fribourg botanical garden. The tropical green house is The tropical greenhouse is located at point 1
DBGI Methodology
Collection
To collect plants in this greenhouse the DBGI methodology was applied. The collection method consists of cutting one or several leaves (depending on the size), wrap them in a coffee filter and put them in a falcon tube with a perforated cap. Each falcon tube is pre-labelled with a unique code assigned to the extract and use as an identifier throughout the experiments. The tube are then store in liquid nitrogen and finally put in the freezer (-80°C) if they are not freeze-dried immediately. Each collected plant, will be report as a point on a qfield map, with several additional data such as the precise location, identification of the plant if possible and minimum five photos. These photos show :
- 1) The plant identification panel
- 2) The plant identification panel with the labelled falcon tube
- 3) A general photo of the plant
- 4) A detail photo of the plant
- 5) The cutting area with the scalpel
- 6) An additional photo can be taken if necessary
Extraction
All plant extracts are freeze-dried and store in falcon tube with non perforated cap and store in a labelled container.
The first step of the process is to weigh out 50 micrograms of each extract and place it in an Eppendorf 2ml with rounded bottom. We accepted an 5% of the weight as an error (±2.5 micrograms). Then add thee metal beads (4mm) inside the tube which will be used to crush the extracts. The plant remains are returned to the falcon and stored in the container.
 Dry plant ready to be weighed
Dry plant ready to be weighed
The sample are then passed in the Retsch machine (shaker) for 2.5 minutes at 25Hz. In this process, the plants are ground to a fine powder.
 Eppendorf tubes with grounded plants
Eppendorf tubes with grounded plants
Once the plants have been ground, the tubes are carefully reopened and 1.7 ml of the extraction solution is pipetted into each tube. The extraction solution mix is composed of 80% methanol, 20% distilled water and 0.1% formic acid. The tubes are then returned to the Restch machine to be shaken again for 2.5 minutes at 25Hz.They are then centrifuged for 2 minutes at 13'000RPM to separate the supernatant from the plant deposits.
Finally, the supernatant is collected and pipetted into a vials tube with hermetically sealed caps. Approximately 1.4 ml of supernatant can be recovered. These tubes are then stored in a labelled container at -80°C.
 Vials tube containing extracts of various plants from the tropical greenhouse
Vials tube containing extracts of various plants from the tropical greenhouse
Analysis
120 microliters of plant extracts are taken from the vials to form aliquots into new vials containing an insert. Those new vials are closed with a slipped cap. These aliquots are analyzed by liquid chromatography mass spectrometry. Spectra are then analyzed using bioinformatics tools.
 Aliquots ready for the mass spectrometry
Aliquots ready for the mass spectrometry
Inaturalist resources
All photos taken at the time of collection are then upload on iNaturalist platform. This help us to check the identification of the plant by external experts and enables us to correct our identifications if necessary. It also benefits iNaturalist, as some plants not included in the database have been added by our request
Here a link to the list of the plants collected in the tropical greenhouse https://www.inaturalist.org/observations?nelat=46.793262993305206&nelng=7.155081027961212&page=2&place_id=any&subview=map&swlat=46.79316199515332&swlng=7.154816830373245&user_id=dbgi&verifiable=any
Sample tracking
Because of the large number of samples, it is important to have reliable sample tracking. As said before, as soon as the samples are collected, each plant is assigned a unique code. This code will follow the plant through each stage of extraction, analysis and storage. This unique code will be report on each different tube during the extraction process and all data relating to this sample are added to the database. By consulting this database, we can quickly find out which plant has already been extracted, how many times it has been extracted, the exact weight that have been extracted and in which container the various tubes are stored. The whole process is automated, and the database automatically updated.
Transitioning from DBGI to EMI
Background
The problematic
In this section, we will take a closer look at a transition project between DBGI and EMI. Until now, the methods implemented were adapted to the collection, extraction and profiling of plants from the botanical garden. However, it is important to adapt this methodology in order to broaden our possibilities and move closer to a solid methodology for the EMI project. There are several aspects that complicate the method in uncontrolled environments. Firstly, unlike in a botanical garden, plants and other organisms are not labelled in a wild environment. This makes the use of the Inaturalist platform as a control by others all the more important. Sufficient good-quality photos should be taken of each sample. On the other hand, the collection of new types of samples implies the implementation of new collection and extraction methodologies. And finally, data processing methods also need to be updated, as we take samples containing many different organisms. The links between these need to be established in the data processing.
A cultivated field is an ideal place to start the transition. The composition of the crops is known, and a certain biodiversity is present within the field. Other organisms such as insects and soil macrofauna can also be collected quite easily.
The DOK trial
Historical background
In the 1970s, discussions on food production in relation to the environment and yield had already been generating debate for several decades. Some pioneers of organic farming practice and scientific from the Research Institute of Organic Agriculture (FiBL) and from the Swiss federal research center for agricultural chemistry and environmental hygiene founded the DOK trial in 1978.
The DOK trial is a long term experiment comparing three cropping systems. DOK stands for BioDynamic, bioOrganic and Konventionell. These three farming system have been studied for 45 years. The DOK trial mimics organic and conventional certified farming methods that are practiced in Switzerland. Since the start of the trial, a large number of scientific publications have been produced thanks to this long-term experiment. Important topics such as climate adaptation, biodiversity, soil quality and yield have been studied on this site.
The site
The DOK site is located in the canton of Baselland, halfway between Biel-Benken and Therwil. The area is located in the Rhine Valley. The floor of this Valley are filled with a layer of gravel and covered by loess material (fine Calcareous silt). This large layer of1 to 1.3 m deep created a fertile soil, that allows water to rise through capillary transport during the summer. The climate is favorable for agriculture, with temperatures averaging 9.7° C until the end of the last century and current annual rainfall of 872 mm. However, average temperatures have been rising steadily in recent decades, with an average of 11.2°C between 2010 and 2020. Climate change and crop resilience are also being studied at DOK, with a view to future extreme weather events (drought, heavy rain, hail, etc.).
Experimental design
The field is divided into 96 plots of 5x20 m. A total of 8 different treatments are applied. Both biodynamic (BIODYN) and bioorganic (BIOORG) systems receive slurry and farmyard manure. The conventional system (CONFYM) also receive slurry and farmyard manure but additionaly it is also treated with mineral fertilizers. This three treatment are applied with two different intensities : 0.7 and 1.4 LU per hectare. A second conventional treatment (CONMIN) system is only treated with mineral fertilizer. In addition, there is control treatment (NOFERT) without any fertilizer. Manure treatment differs between systems, and consequently organic carbon and nitrogen inputs also differ. The conventional system (CONFYM) uses stacked manure, the bioorganic system (BIOORG) uses rotted manure and the biodynamic system (BIODYN) uses compost manure. These are aerated more intensively and for longer, respectively. Mineral fertilizers comply with Swiss Federation regulations for concentrations of calcium-ammonium-nitrate, urea, potassium chloride and triple super phosphate.
Plant protection in both conventional systems consists of the application of herbicides, fungicides, insecticides, and molluscicides. In BIOORG and BIODYN the plant protection follows the standards (Lampking 1990). In addition, the BIOORG system treats potatoes with copper sulfate (CuSO4). The Bacillus thuringiensis bacterium is sprayed against potato beetles in BIOORG and BIODYN systems. The NOFERT system uses the same protections as the organic systems.
3 crops are planted simultaneously (columns A, B and C in the schema) and a 7-year crop rotation is carried out in the plot following this sequence since 2013 : Maize, Soybeans, Vinter wheat 1 + catch crop, Potatoes, Winter Wheat 2, Grass-clover 1, Grass-clover 2. Grass-clover mixture is composed of Trifolium pratense 6%, Trifolium repens 12%, Dactylis glomerata 17%, Festuca pratensis 36%, Lolium perenne 21% and Phleum pratense 12%.
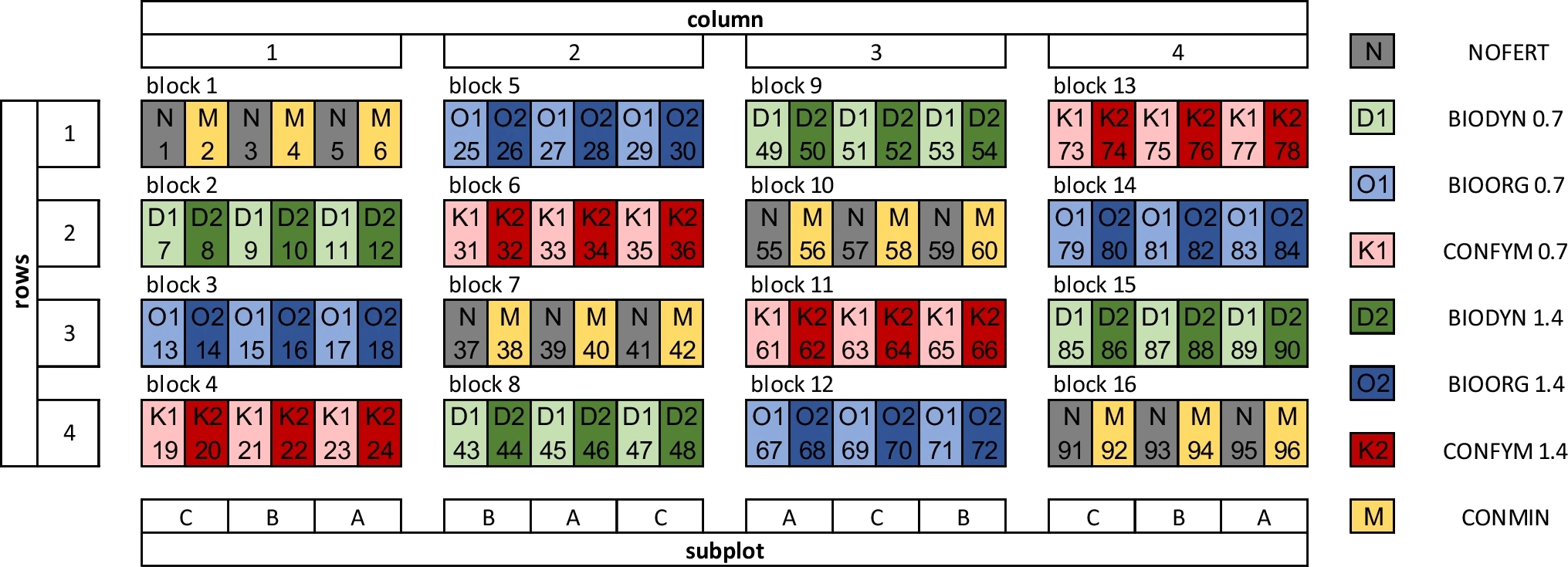 Field setup of the DOK trial with spatial orientation of blocks, columns, rows, and subplots. Each year three crops are cultivated in the subplots A, B, and C. N, NOFERT, unfertilized; D, BIODYN, biodynamic; O, BIOORG, bioorganic; K, CONFYM, conventional with farmyard manure; and M, CONMIN, conventional purely mineral fertilization. Organic fertilization: 0.7 and 1.4 correspond to organic fertilization at 0.7 and 1.4 livestock units per hectare. (H-M Krause et al. 2022)
Field setup of the DOK trial with spatial orientation of blocks, columns, rows, and subplots. Each year three crops are cultivated in the subplots A, B, and C. N, NOFERT, unfertilized; D, BIODYN, biodynamic; O, BIOORG, bioorganic; K, CONFYM, conventional with farmyard manure; and M, CONMIN, conventional purely mineral fertilization. Organic fertilization: 0.7 and 1.4 correspond to organic fertilization at 0.7 and 1.4 livestock units per hectare. (H-M Krause et al. 2022)
Previous experiments and results obtained
Around 200 studies based on the DOK trial have been published to date. Some interesting results from different major studies conducted on the DOK trial site :
On average, organic crop yields are about 20% lower (https://doi.org/10.7717/peerj.15000) (although yields of specific crops vary) compared to the conventional system receiving mineral fertilizers, while nutrient inputs in the conventional system are 60% higher for mineral nitrogen, 45% for potassium and 40% for phosphorus (https://www.cabidigitallibrary.org/doi/full/10.5555/20063115035).
Organic systems reduce fertilizer and energy inputs by 34 to 53% and pesticides by 97%. An improvement in soil fertility and biodiversity is observed in organic system plots. These plots are less dependent on external inputs. (https://doi.org/10.7717/peerj.15000)
The soil organic matter (humus) content remained stable only in the biodynamic system during the first 21 years of the trial. All other systems showed a decline in humus. This difference is significant between the biodynamic and the purely mineral conventional systems. The soil microbial biomass showed 25% lower values in the BIODYN and CONFYM while system without manure application (CONMIN) showed 34% lower values. (https://doi.org/10.1016/j.agee.2006.05.022)
In term of nutrition quality, a broad spectrum of molecules relevant to human nutrition have been identified in wheat including analysis of : seed weight, protein content, phosphate levels, antioxidative capacity of phenols, fiber, fructan, oxalate and phytic acid. A clear difference in grain weights from the different farming systems was observed. In fact, thousand-seed weight and protein were significantly lower in unfertilized wheat. On the contrary, total oxalate was significantly higher in these plots. For the rest, and for the majority of substances of nutritional interest, no significant differences were observed between biodynamic, organic and conventional farming systems. (https://core.ac.uk/outputs/235697697/?utm_source=pdf&utm_medium=banner&utm_campaign=pdf-decoration-v1)
In terms of energy efficiency, organic crops require 30-50% less energy to produce per unit area. This includes the energy needed to produce fertilizers and pesticides. Taking into account the higher yield per unit area in conventional systems, the energy used per unit yield is still 19% lower for organic farming system.
Long-term measurements show a nitrogen deficit in all cultural systems. This means that the cultivated plants used more nitrogen than what was provided to them by the fertilizer applied. This additional nitrogen comes from soil mineralization, nitrogen fixation by legumes and their symbiosis, and aerial deposition. Regarding the phosphorus and the potassium, the deficits are much greater in biological and biodynamic systems than in conventional systems.
https://www.fibl.org/en/locations/switzerland/departments/soil-sciences/bw-projekte/dok-trial
DOK project experiments
Global experimental design
For our experiments, we decide to compare the BIODYN and the CONMIN systems which are probably the two most different systems in terms of cultivation methods. 4 experiments were design, that have been replied 4 times (one in each replicate plots). The aim was to see if there is any metabolomic differences in the crops (Wheat) and the other organisms present around the culture such as weeds and invertebrates (insects, mollusks, annelids and other soil macrofauna). Crop A was used in all 4 experiment, which this year was the montalbano variety of wheat. Our experiments were carried out in plots with the following numbers: 6, 40, 56 and 96 for CONMIN plots and 12, 46, 50 and 90 for BIODYN plots.
Collection review
The collection took place on April 18 from 1:30 p.m. to around 7 p.m. at the DOK trial (Localization : 47.502578, 7.539575 (Therwil, Basel canton)). The collection went smoothly, with cool weather. It didn't rain in the afternoon, but conditions were damp because it had rained that very morning. We didn't find many insects on site, probably because the weather was too cold for them, but we did collect a few, along with slugs and soil macrofauna. All the wheat fields (with no difference between cropping systems) had been infected by a fungal disease, which may have an influence on the metabolomic results.
Collection methodology
Equipment list for one afternoon collection
- 80 Falcon tubes with label
- 80 Falcon tubes (reserve without label)
- 80 Label (reserve)
- 16 big zip lock bag
- 1/pers tablet / smartphone with Qfield application and the project synchronized
- 1/pers scalpel + a spare in case
- Gloves : 5 pairs/pers (each size)
- Liquid nitrogen
- Thermos for the liquid nitrogen
- Shovels
- Measuring tape
- Big boxes
- 1/pers Ethanol 70% spray
- Sardines
- Rope / string
- External battery with cables for each type of device
- Indicator arrows
- 160 Coffee filter
- Scissors
- Sharp knife
- Soil corer
The following paragraphs presents the 4 experiments carried out on the site, with the aim of testing different collection and analysis methods.
20x20 crushed : chemodiversity
Experimental concept
We randomly select a 20cm x 20cm square in the plot, if possible avoiding edge effects. We cut everything in this square above ground level. The cultivated plant as well as any weeds growing in between and possibly insects, invertebrate or mushroom that were present in the area. All this samples are store in a Ziplock with an unique code. They will then be extracted as one entities to rate and qualify the chemodiversity of the parcelle.
Methodology
We applied the methodology once in each of the 8 plots. Each type of agriculture is replicated four times on the DOK trial. That is allowing us to have four replications of above-ground-biodynamic specimens and four replication of above-ground-conventional specimens.
Collection
The 20cm X 20cm square is draw on the ground using a measuring tape. The location is randomly chose in on of the side row of the plot (the middle row is reserved for measuring yield and cannot be touched). All the crops, weeds and others which are found in this square are cut at their base and put in zip-lock prelabeled with an unique code stick on the inside, as it falls off easily when exposed to the cold. The entire zip-lock is then immersed in liquid nitrogen. The sample are store at -80°C waiting for the extraction process.
During the collect the zip-lock partially destroyed in liquid nitrogen. after further test we dicide that paper bag should be use for the liquid nitrogen and the dry-freezer for the next collect. But zip lock are indispensable after the lyophilization to keep the sample dry.
Extraction
s
Result
Emi_id of the sample and directus link (lab extract)
This experiment groups samples with the following identity:
- fibl_000081 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000081_01
- fibl_000084 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000084_01
- fibl_000086 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000086_01
- fibl_000088 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000088_01
- fibl_000090 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000090_01
- fibl_000092 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000092_01
- fibl_000094 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000094_01
- fibl_000096 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000096_01
Note that the link to directus are only available with a directus account and using the vpn of the University of Fribourg.
Analysis of the statistical data and spectra is still in progress.
Mesofauna and Macrofauna extraction
Experimental concept
Compare the mesofauna of the soil between the conventional and the biodynamic agriculture. We decide to use the Berlese extractor to collect the mesofauna. We want to extract the metabolite of the entire sample without sorting the invertebrate and use this to rate and qualify the chemodiversity of the soil mesofauna. The megafauna (earthworm, centipede, spiders...) is sorted out before the Berlese extraction and analyzed as individual extracts.
The experiment is replicated four times for each type of agriculture systems. The soil block is collected in a random place in the plot (avoiding the center line, which cannot be touched).
Methodology
Collection
A place is randomly chose in the side row of the 5m x 20m plot. To limit the crop damage, we took the soil sample right under the 20cm x 20cm cut for the previous method. A bloc of soil of 10cm x 10cm and 5cm deep is collected using a flat shovel and store in a zip-lock prelabelled with an unique code stick on the inside. The block is collected in such a way as to maintain the integrity of the soil structure as much as possible. The soil blocks are store in a shaded area until the end of the collection process. They are then store in the 4°C fridge to await the extraction.
Berlese Extraction
First of all, the block of earth is gently disassembled by hand. The megafauna is set aside. Each individual is placed in a pre-labelled falcon tube and an individual observation is added to qfield (located where the soil was taken). The observation is done exactly the same as the observation of plants. Photos are taken, localization, and, if known, the species name (If not, "unknow" will be filled in the box).
The megafauna falcon tubes are store in the -80°C and then dry-freezed during 72 hours. Those specimens will then be extracted with the dbgi process.
The mesofauna is extracted by the Berlese extractor. The soil is placed in a sieve and gently compacted. The sieve is placed over a funnel and a container of ethanol is placed directly underneath. A bulb that heats up is placed above the earth and this heat dries the soil from above. The mesofauna try to escape the heat and drought by escaping from below, eventually falling into the ethanol.
 Installing of two Berlese extractors
Installing of two Berlese extractors
Extraction (after Berlese)
Photos of the mesofauna are done to future identification of the specimens with the binocular loupe. Then the falcon tube are centrifuged for 5 min at 5000 rps. The pellet is removed using a pipette with a cut tip and conserve in a eppendorf tube already weighted. If necessary, centrifuge the Eppendorf tube for 2 min at 13'000 rps, take out the supernatant and put the end of the pellet (if too much for). Remove the supernatant in the Eppendorft tube, after centrifugation (2 min at 13'000 rps) and dry the extract with sample concentrator machine.
Once dry, the Eppendorft tube are weighted again and the mass of the specimens is add to the database. Then, a metal bead is placed on the tube and the further extraction process is done as dbgi extraction process.
Additionally 1700 microliter of the ethanol of each sample is collected and put in a vial tube to see if any molecule have been extracted during the Berlese processus.
Result
Emi_id of the sample and directus link (lab extract)
This experiment groups samples with the following identity:
- fibl_000083 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000083_02
- fibl_000085 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000085_02
- fibl_000087 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000087_02
- fibl_000089 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000089_02
- fibl_000091 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000091_02
- fibl_000093 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000093_02
- fibl_000095 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000095_02
- fibl_000097 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000097_02
The link to directus are only available with a directus account and using the vpn of the University of Fribourg.
Analysis of the statistical data and spectra is still in progress. The data from these analyses will probably be irrelevant, as many of the samples were lost during the various laboratory stages. Part of the mesofauna dried inside the funnel during the Berlese extraction. Some of the samples were blown out of the tubes during drying with the sample concentrator.
Photos and Inaturalist Id
Here, some photos taken of our samples with the binocular loupe.
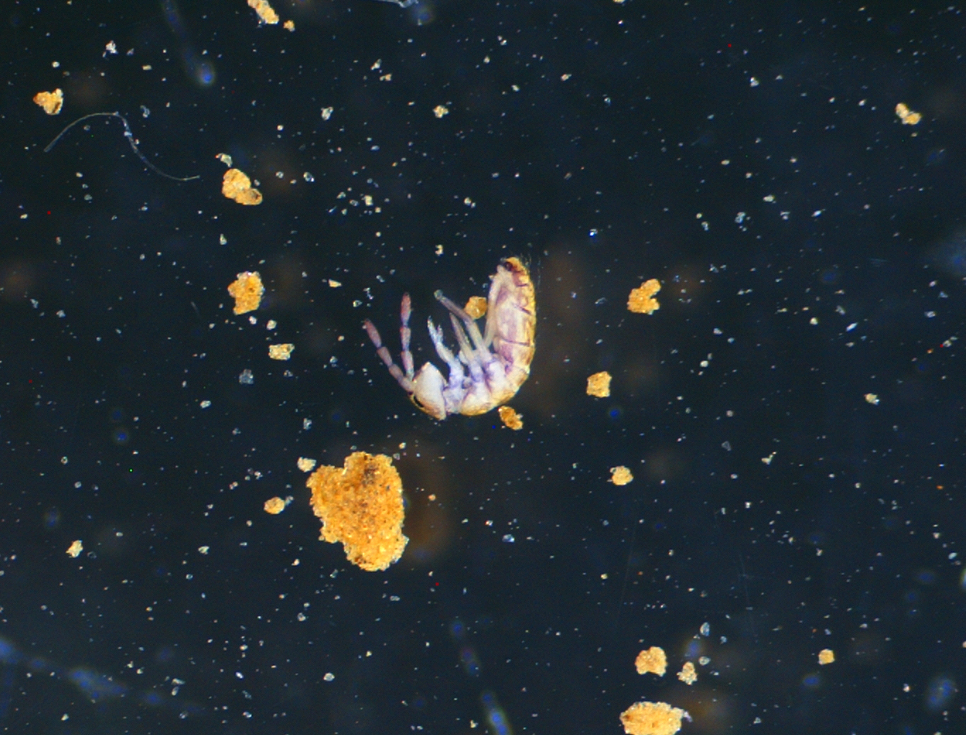 Specimen from the fibl_000087 batch. This have been identified as a member of the Isotomoidea Superfamily (Collembola subclass) by a member of Inaturalist community. Inaturalist link with other observations of the same species/genera/family in our samples: https://www.inaturalist.org/observations/216301081
Specimen from the fibl_000087 batch. This have been identified as a member of the Isotomoidea Superfamily (Collembola subclass) by a member of Inaturalist community. Inaturalist link with other observations of the same species/genera/family in our samples: https://www.inaturalist.org/observations/216301081
 Specimen from the fibl_000091 batch. This have been identified as a member of acari subclass by a member of Inaturalist community. Inaturalist link with other observations of the same species/genera/family in our samples : https://www.inaturalist.org/observations/216300230
Specimen from the fibl_000091 batch. This have been identified as a member of acari subclass by a member of Inaturalist community. Inaturalist link with other observations of the same species/genera/family in our samples : https://www.inaturalist.org/observations/216300230
 Specimen from the fibl_000085 batch. This have been identified as an hymenoptera by a member of the Inaturalist community. Inaturalist link of the observation : https://www.inaturalist.org/observations/216289778
Specimen from the fibl_000085 batch. This have been identified as an hymenoptera by a member of the Inaturalist community. Inaturalist link of the observation : https://www.inaturalist.org/observations/216289778
 Specimen from the fibl_000087 batch. This have been identified as a member of the Geophilidae family (Chilopoda class) by a member of Inaturalist community. Inaturalist link with other observations of the same species/genera/family in our samples : https://www.inaturalist.org/observations/216299547
Specimen from the fibl_000087 batch. This have been identified as a member of the Geophilidae family (Chilopoda class) by a member of Inaturalist community. Inaturalist link with other observations of the same species/genera/family in our samples : https://www.inaturalist.org/observations/216299547
 Specimen from the fibl_000087 batch. This have been identified as a member of the Onychiuridae family by a member of Inaturalist community. Inaturalist link of other observation of the same species/genera/family in our samples : https://www.inaturalist.org/observations/216287636
Specimen from the fibl_000087 batch. This have been identified as a member of the Onychiuridae family by a member of Inaturalist community. Inaturalist link of other observation of the same species/genera/family in our samples : https://www.inaturalist.org/observations/216287636
 Specimen from the fibl_000087 batch. This have been identified as a member of the Lithobiomorpha order (Chilopoda class) by a member of the Inaturalist community. Inaturalist link of the observation : https://www.deepl.com/translator#fr/en/ordre%20Lithobiomorpha
Specimen from the fibl_000087 batch. This have been identified as a member of the Lithobiomorpha order (Chilopoda class) by a member of the Inaturalist community. Inaturalist link of the observation : https://www.deepl.com/translator#fr/en/ordre%20Lithobiomorpha
More photos of specimens can be seen on the following inaturalist page: https://www.inaturalist.org/observations?page=2&place_id=any&user_id=dbgi&verifiable=any
Plants pair comparison
Experimental concept
Found some plants that are present in the biodynamic plot and the conventional mineral one and compare their metabolomic content.
Methodology
The first step is to make a brief tour of the fields and a list of the species present in the two plots (also list unknown species). For this experiment, we found three species present in both cropping systems:
- The main crop: Triticum aestivum of the Montalbano variety
- The remains of last year's crop: Solanum tuberosum
- A species of poaceae
Collection and extraction
Collections and extractions are carried out using the same methodology as for the DBGI.
Result
Emi_id of the sample, directus link (lab extract) and Inaturalist link
This experiment groups samples with the following identity:
Triticum aestivum:
- fibl_000007 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000007_01 / https://www.inaturalist.org/observations/208862402
- fibl_000010 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000010_01 / https://www.inaturalist.org/observations/208863232
- fibl_000014 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000014_01 / https://www.inaturalist.org/observations/208862416
- fibl_000015 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000015_01 / https://www.inaturalist.org/observations/208863250
- fibl_000019 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000019_01 / https://www.inaturalist.org/observations/208862436
- fibl_000020 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000020_01 / https://www.inaturalist.org/observations/208862445
- fibl_000021 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000021_01 / https://www.inaturalist.org/observations/208862463
- fibl_000022 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000022_01 / https://www.inaturalist.org/observations/208862481
- fibl_000025 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000025_01 / https://www.inaturalist.org/observations/208863268
- fibl_000028 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000028_01 / https://www.inaturalist.org/observations/208862500
- fibl_000032 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000032_01 / https://www.inaturalist.org/observations/208862514
- fibl_000037 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000037_01 / https://www.inaturalist.org/observations/208862543
- fibl_000038 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000038_01 / https://www.inaturalist.org/observations/208862562
- fibl_000044 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000044_01 / https://www.inaturalist.org/observations/208862580
- fibl_000046 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000046_01 / https://www.inaturalist.org/observations/208862592
- fibl_000051 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000051_01 / https://www.inaturalist.org/observations/208862607
- fibl_000053 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000053_01 / https://www.inaturalist.org/observations/208862628
- fibl_000054 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000054_01 / https://www.inaturalist.org/observations/208862641
- fibl_000056 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000056_01 / https://www.inaturalist.org/observations/208862660
- fibl_000058 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000058_01 / https://www.inaturalist.org/observations/208862697
- fibl_000059 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000059_01 / https://www.inaturalist.org/observations/208862719
- fibl_000065 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000065_01 / https://www.inaturalist.org/observations/208863283
- fibl_000069 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000069_01 / https://www.inaturalist.org/observations/208863308
- fibl_000073 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000073_01 / https://www.inaturalist.org/observations/208863337
- fibl_000074 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000074_01 / https://www.inaturalist.org/observations/208863352
Solanum tuberosum :
- fibl_000003 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000003_01 / https://www.inaturalist.org/observations/208862738
- fibl_000006 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000006_01 / https://www.inaturalist.org/observations/208862284
- fibl_000008 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000008_01 / https://www.inaturalist.org/observations/208862299
- fibl_000027 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000027_01 / https://www.inaturalist.org/observations/208862333
- fibl_000030 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000030_01 / https://www.inaturalist.org/observations/208863120
- fibl_000031 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000031_01 / https://www.inaturalist.org/observations/208863143
- fibl_000035 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000035_01 / https://www.inaturalist.org/observations/208862346
- fibl_000036 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000036_01 / https://www.inaturalist.org/observations/208862360
- fibl_000043 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000043_01 / https://www.inaturalist.org/observations/208863157
- fibl_000047 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000047_01 / https://www.inaturalist.org/observations/208862374
- fibl_000061 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000061_01 / https://www.inaturalist.org/observations/208863170
- fibl_000064 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000064_01 / https://www.inaturalist.org/observations/208863433
- fibl_000066 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000066_01 / https://www.inaturalist.org/observations/208863184
- fibl_000076 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000076_01 / https://www.inaturalist.org/observations/208863207
Poaceae
- fibl_000004 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000004_01 / https://www.inaturalist.org/observations/208862937
- fibl_000018 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000018_01 / https://www.inaturalist.org/observations/208862966
- fibl_000026 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000026_01 / https://www.inaturalist.org/observations/208862980
- fibl_000039 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000039_01 / https://www.inaturalist.org/observations/208863000
- fibl_000048 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000048_01 / https://www.inaturalist.org/observations/208863023
- fibl_000068 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000068_01 / https://www.inaturalist.org/observations/208863037
- fibl_000070 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000070_01 / https://www.inaturalist.org/observations/208863063
- fibl_000080 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000080_01 / https://www.inaturalist.org/observations/208863097
The link to directus are only available with a directus account and using the vpn of the University of Fribourg.
Soil extraction
Experimental concept
Collection of soil from all plots and analysis of its metabolomic content. The soil is sampled using a small core.
Methodology
Collection and extraction
Soil is taken at random from each of the 8 plots (4 replications of each cropping system). To limit damage to the field, we sample the soil in the same place as the soil block was sampled for the Berlese extraction. The soil core is then placed in a caffeine filter and immersed in liquid nitrogen. Further extraction is carried out using the DBGI methodology.
Result
Emi_id of the sample and directus link (lab extract)
This experiment groups samples with the following identity:
- fibl_000005 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000005_01
- fibl_000011 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000011_01
- fibl_000016 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000016_01
- fibl_000024 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000024_01
- fibl_000033 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000033_01
- fibl_000042 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000042_01
- fibl_000050 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000050_01
- fibl_000055 : http://directus.dbgi.org/admin/content/Lab_Extracts/fibl_000055_01
The link to directus are only available with a directus account and using the vpn of the University of Fribourg.
Metabolomics analysis
Methodology
LC
MS
Data treatment
Result
Untargeted analysis
The results of unsupervised statistics show a clustering by taxonomy. This show us that the biggest difference in term of metabolomic content is coming from the difference in between the species.
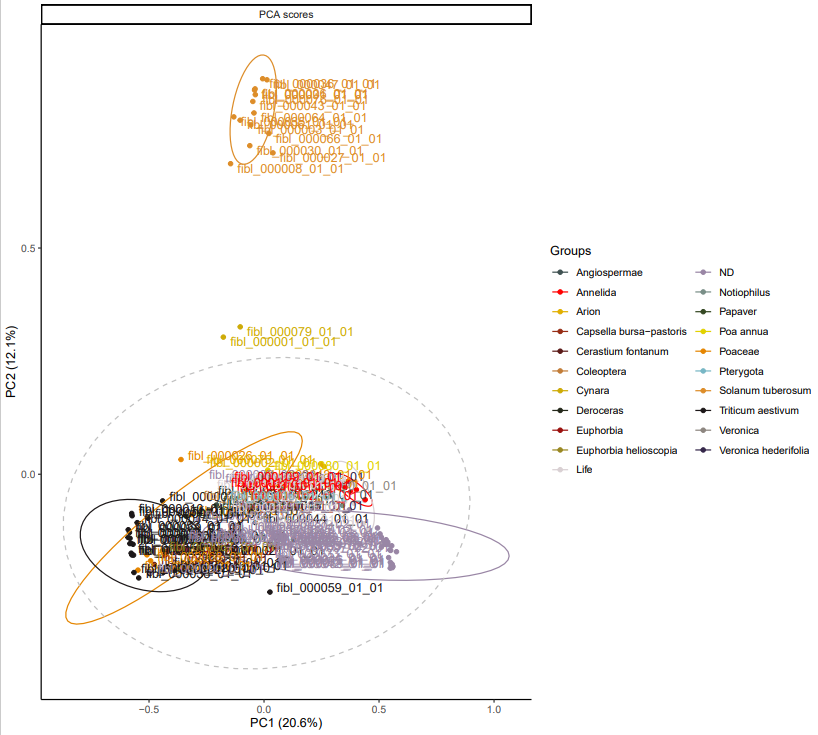
*PCA for dataset fibl_pilot_pos. Comparison across all taxa.
The unsupervised results for wheat alone do not show clustering by cropping system either.
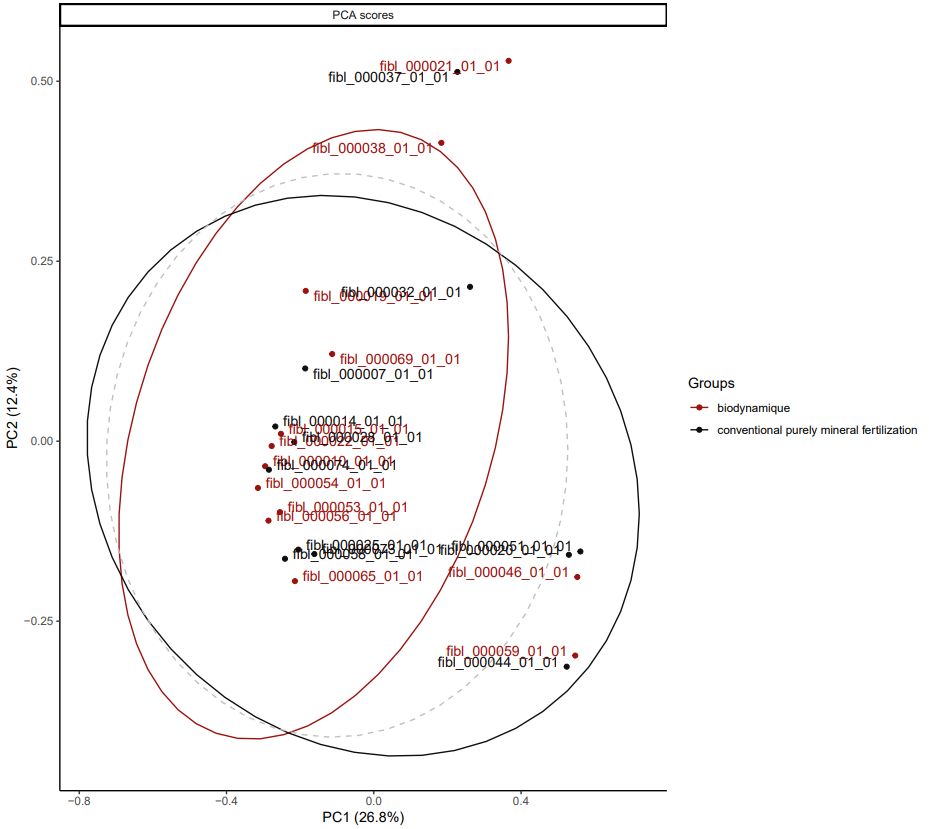 *PCA for dataset fiblpilot_pos. Comparison across biodynamic vs conventional purely mineral fertilization. Filter taxon _Triticum aestivum only.
*PCA for dataset fiblpilot_pos. Comparison across biodynamic vs conventional purely mineral fertilization. Filter taxon _Triticum aestivum only.
However, for Solanum tuberosum a difference is already visible in the unsupervised statistics. Two distinct clusters are formed according to cultivation methods.
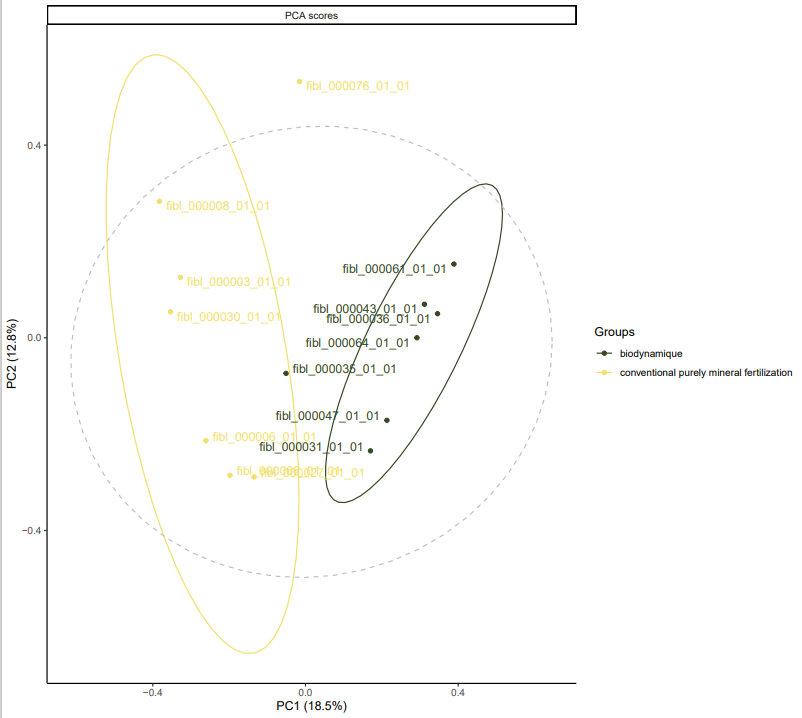 *PCA for dataset fiblpilot_pos. Comparison across method biodynamic vs conventional purely mineral fertilization. Filter taxon _Solanum tuberosum . Filter Variable Metadata Status : above sirius confidencescore (0.5).
*PCA for dataset fiblpilot_pos. Comparison across method biodynamic vs conventional purely mineral fertilization. Filter taxon _Solanum tuberosum . Filter Variable Metadata Status : above sirius confidencescore (0.5).
Supervised analysis
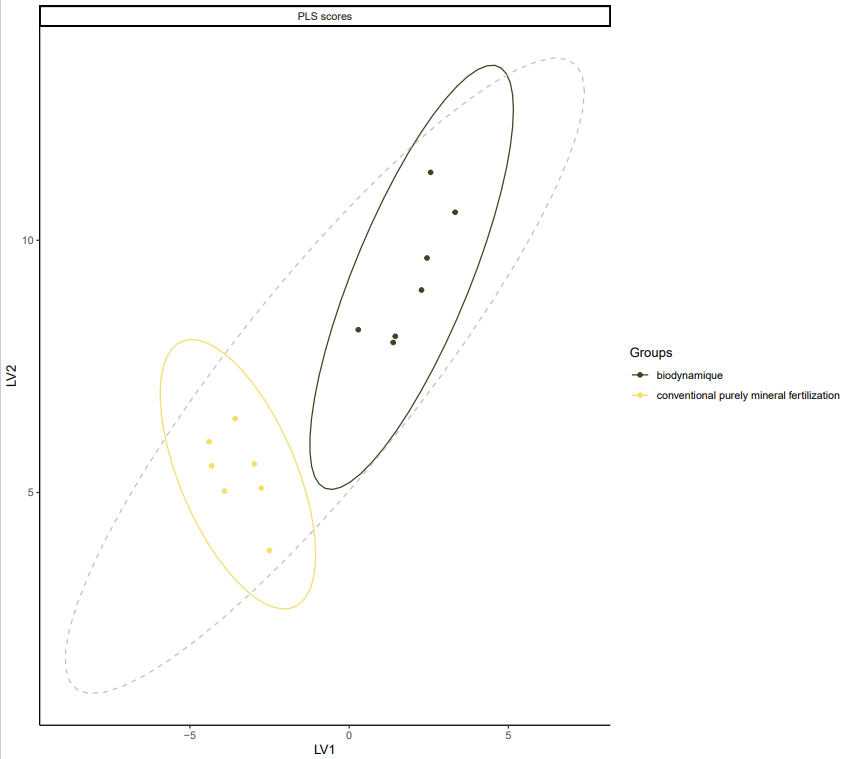
Supervised statistic, with maximizing the difference based on the farming system, a clustering by BIODYN and CONMIN appears in the wheat samples analysis. The metabolomic content is not the same in the wheat crops that are growing in different farming system.
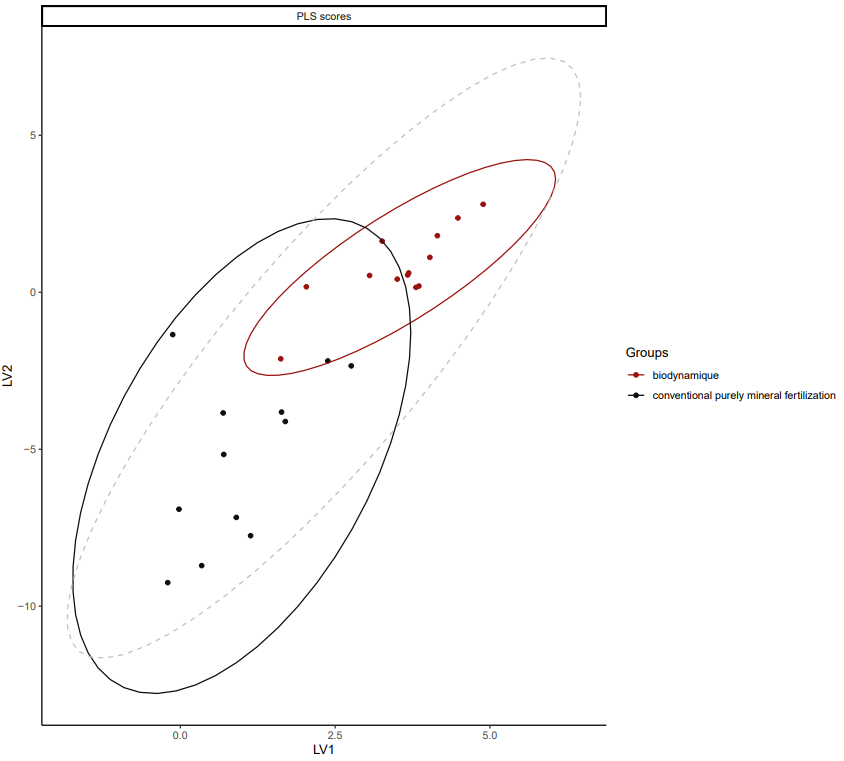 *PLSDA for dataset fiblpilot_pos. Comparison across method biodynamic vs conventional purely mineral fertilization. Filter taxon _Triticum aestivum. Filter Variable Metadata Status : above sirius confidencescore (0.5).
*PLSDA for dataset fiblpilot_pos. Comparison across method biodynamic vs conventional purely mineral fertilization. Filter taxon _Triticum aestivum. Filter Variable Metadata Status : above sirius confidencescore (0.5).
By looking the molecules that are causing this difference in the wheat crop we can spot some endogen molecules such as pipecolic acid and also exogenous molecules such as the metribuzin which is a well know herbicide. Both of this molecules are present in the CONMIN system. First molecule of importance is the pipecolic acid that is required for the systemic acquired resistance in plant and overrepresented in the CONMIN farming system.
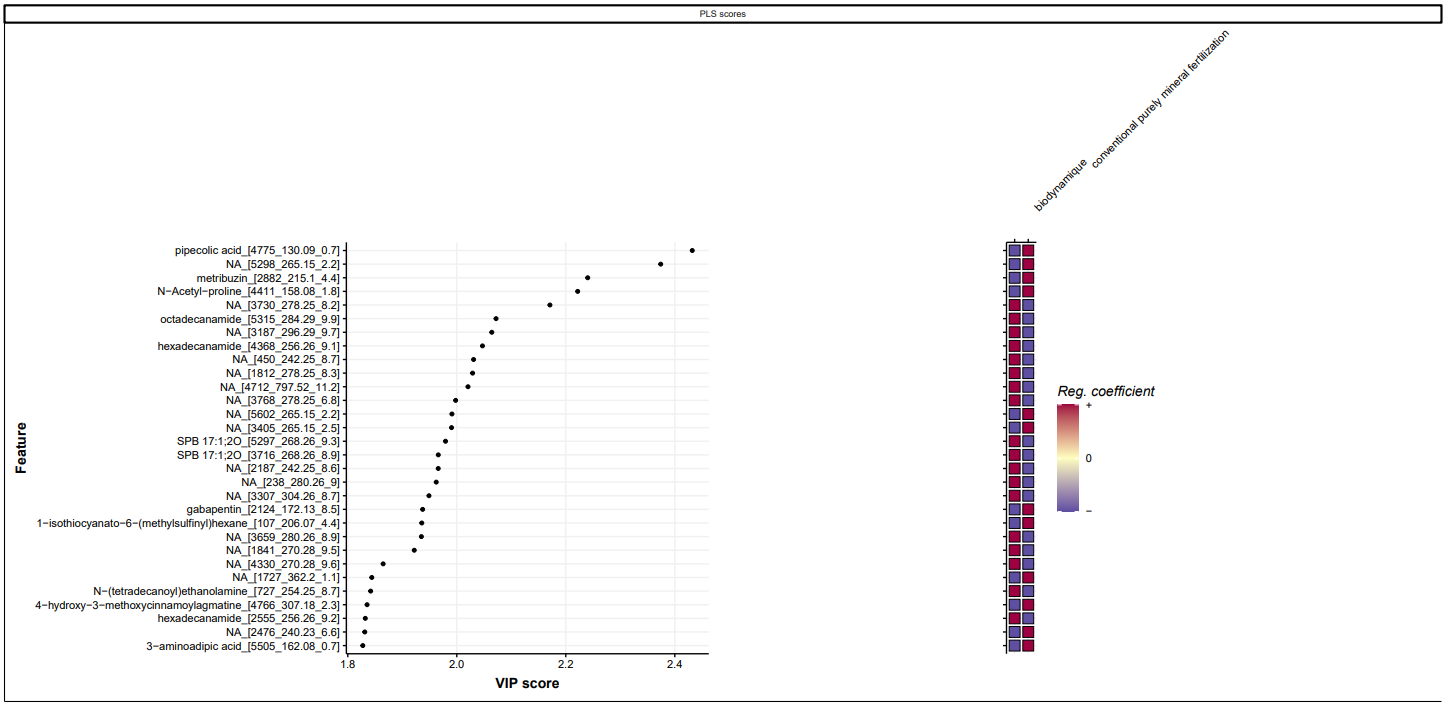 *PLSDA selected Features of Importance for dataset method biodynamic vs conventional purely mineral fertilization above sirius confidencescore scaled.
*PLSDA selected Features of Importance for dataset method biodynamic vs conventional purely mineral fertilization above sirius confidencescore scaled.
Although the differences were already visible with the unsupervised statistics, we can also observe a clear clustering of Solanum tuberosum by farming system in the supervised analysis.
 *PLSDA for dataset fiblpilot_pos. Comparison across method biodynamic vs conventional purely mineral fertilization. Filter taxon _Solanum tuberosum . Filter Variable Metadata Status : above sirius confidencescore (0.5). Scaled graph.
*PLSDA for dataset fiblpilot_pos. Comparison across method biodynamic vs conventional purely mineral fertilization. Filter taxon _Solanum tuberosum . Filter Variable Metadata Status : above sirius confidencescore (0.5). Scaled graph.
The naringenin is the first component of importance in the PLSDA of potatoes analysis and is overrepresented in the BIODYN system. This flavonoid is an antioxidant molecule know to have good effect on human health (Venkateswara Rao P, SDVS Kiran, Rohini P and Bhagyasree P. Flavonoid: A review on Naringenin. J Pharmacogn Phytochem 2017;6(5):2778-2783.) Other studies present naringenin as a molecule capable of activating certain resistance genes against pathogens in certain plants (https://doi.org/10.3389/fpls.2021.672552).
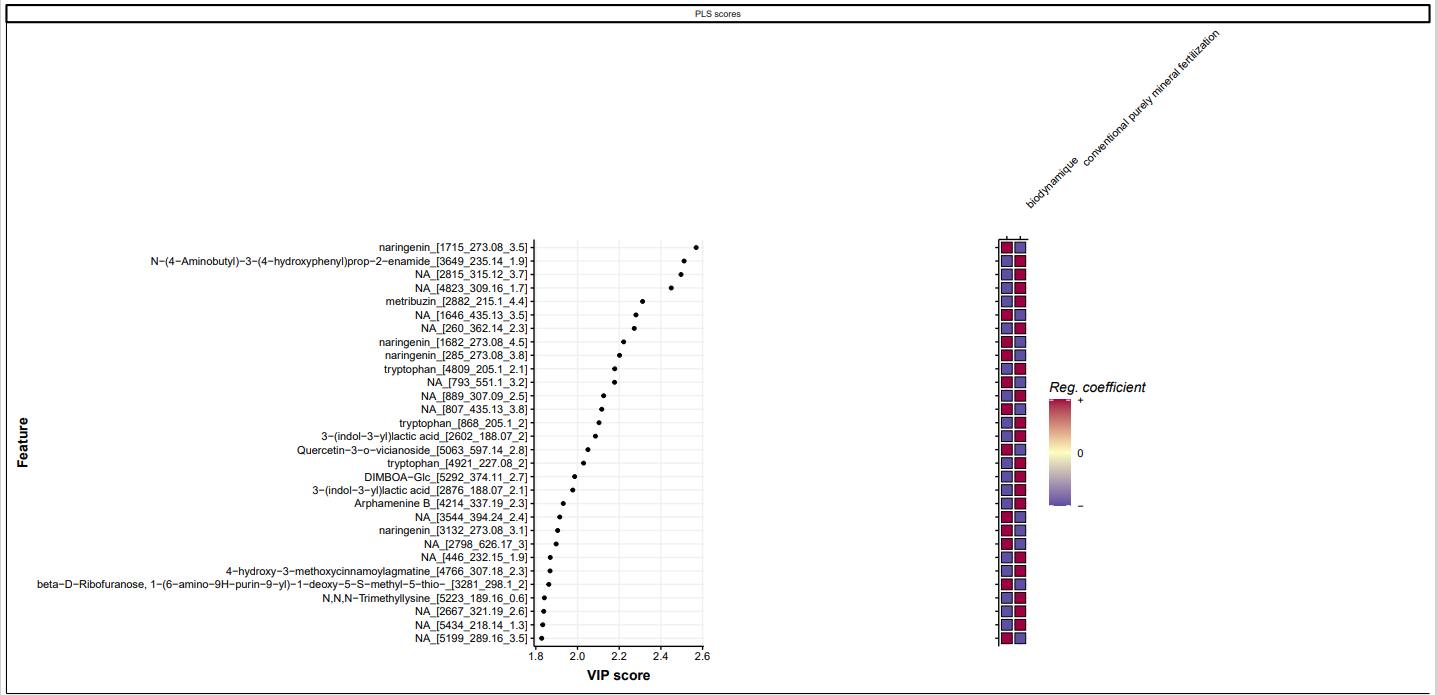 *PLSDA selected Features of Importance for dataset method biodynamic vs conventional purely mineral fertilization above sirius confidencescore scaled.
*PLSDA selected Features of Importance for dataset method biodynamic vs conventional purely mineral fertilization above sirius confidencescore scaled.
Treemap plots show that a big family of hydroxycinnamic acid amides (HCAA) are overrepresented in the CONMIN system. This HCAA molecules are present in both wheats and potatoes crops from CONMIN system. This family is known as a stress induce molecule family associated with plant defense mechanisms (doi: 10.3389/fpls.2022.922119).
The metribuzin is also visible in both treemaps (wheat and potato) in the pseudoalkaloids group.
Potato crops show a strong overrepresentation of flavonoids in the BIODYN system. This is consistent with our molecules of importance, including naringenin and its over-representation in BIODYN plots.
Tree map image
Molecular networking
Around 5600 spectra have been found and are represented in the following molecular network.
Discussion and conclusion
These different experiments showed a metabolomic difference between wheat and potatoes from two different farming systems. The results of these experiments have yet to be completed. But these results are encouraging for future metabolomic studies on this site.
To date, we have been unable to find any explanation for the presence of these stress molecules in wheat and potatoes from the conventional system. Investigations into the metabolism of these compounds and the culture conditions need to be carried out to learn more. We could detect some synthetic substances in the metabolomic content of both crops from CONMIN system. However, a check with field technicians is necessary to ensure that the herbicide detected is indeed the one applied to the plants.
This comparative study has enabled us to establish other important metadata for sample tracking. Our extraction methodologies could be tested on organisms other than plants and were mostly successful. In addition, some ideas on how to relate our samples to each other have been put forward. For example, how can we link organism extractions from the macrofauna of a certain soil with the overall metabolism of that soil? Lab applications and bioinformatics pipelines are being fine-tuned to overcome this problem.
Other methodological aspects remain to be worked on. Methodological alternatives for harvesting in the field where it is not possible to take some of our materials (such as liquid nitrogen or dry ice) have yet to be worked on. Silica gel drying is possible in certain cases, for plants for example, but cannot be used with insects or other invertebrates for reasons of animal ethics. On-site extraction techniques could be considered to avoid sample degradation during transport and customs clearance problems.
References
question a voir avec PMA
- [x] légende image resultat metabo
- [x] est-ce qu'il faut mettre analyse avec sirius confidence ou l'autre sans ?
- [x] lien pour voir la treemap belle Example
- [ ] previous result mettre plus ou ça va?
- [ ] référence
- [x] réseau moléculaire ? je le trouve pas
- [ ] est-ce que ya des résultats avec l'extraction pour les invertébrés (méthode fonctionne ou pas ?)