2024-01-12
This is PMA's DBGI daily open-notebook.
Today is 2024.01.12
Todo today
Have a look at the DBGI discussion forum
- https://github.com/orgs/digital-botanical-gardens-initiative/discussions
Doing
Anticipated LOTUS
- edges with very different weight
- no true negatives
- no edge weight for the negatives
They are models which are positive
Inductive models means that we cannot use node embedding per se
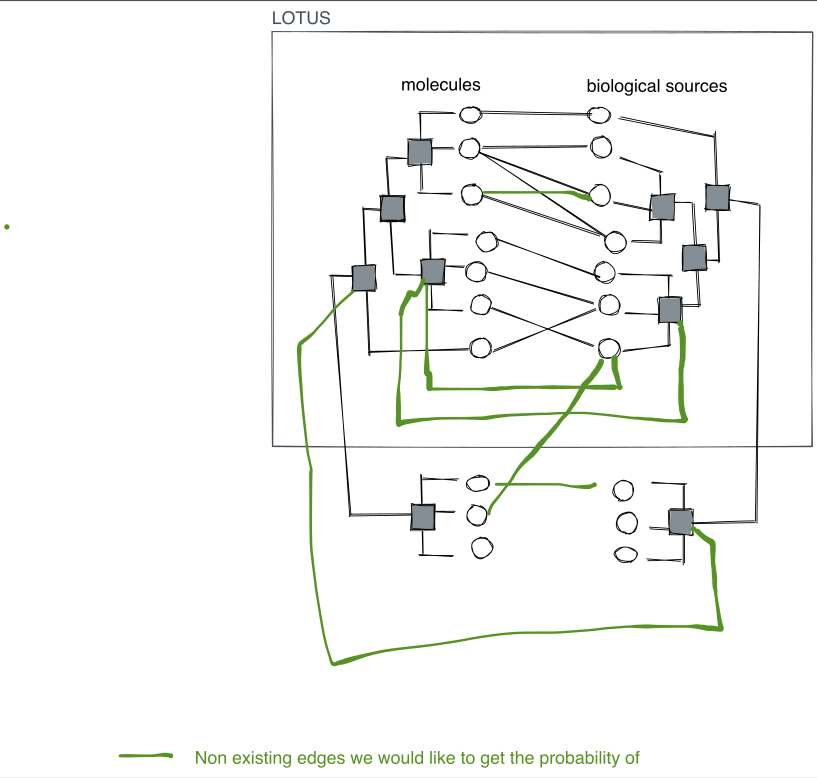
Find common features across biological sources and molecules. The wikidata page of these nodes could be a thing.
SciBERT
Creating a Anticipated Lotus page in the Dendron DBGI vault.
- Textual description at least 10 words of both species and molecules.
This textual description needs also to be as complete as the existing ones. So a WD description would be OK if we keep name, INCHI, SMILES etc, but not the pubchem id for ex.
EMIKG Portal
- Mono repo approach
e.g. google is a mono repo
https://github.com/enpkg/enpkg_full
https://en.wikipedia.org/wiki/Monorepo
Rust monoliths backend : handles the DB, the authorizations, Sample management, etc. No particular reason to do it in Rust except for the security reasons. Should be as integrated as possible.
Rust part listen to API calls from Python modules.
When you expect to have full control of a thing, e.g. you are the provider.
Created emikg-portal
A prefixed tree https://docs.rs/trie-rs/latest/trie_rs/ To integrate.