Notes
Kept below. Not much notes taken. Lot of chat ! We might want to have a different ratio next time.
Starting at 9:30
- Physical at Pavillon Vert. See here for location details https://www.unifr.ch/scimed/fr/pavillonvert
- Remote at https://meet.jit.si/DBGI_talks
Welcome and workshop members’ introduction (15 min)
Present
ACS (Ana Claudia Sima) ED (Emmanuel Defossez) FM (Florence Mehl) HL (Henry Luetcke - remote) LC (Luca Cappelletti) MP (Marco Pagni) MV (Marco Visani) MW (Maëlle Wannier) PMA (Pierre-Marie Allard) TM (Tarcisio Mendes)
intro round
Review: ontology engineering activities (rest of the morning)
-
Organism taxonomy.
-
Relying on WD as an aggregator of taxonomies.
Marco : 2 problems. To have an identifier for a taxon (afterwards not sure why this is not adressed by WD) An identifier with clear release cycles (not so easily tracked at WD) They are WD dumps.
Luca: Keeping the mapping safe is N°1, 2 and 3 problem. No consensus.
BUT, in the meantime, in the DBGI-KG context we will use WD id.
Manu : important to have solution for sampling of soils, water, honeys etc. More generally we should create (or re-use if existing) a statement of this type "macroscopic taxon is / apparently major biological organism of sample is" The biomeaggregation specimen type defined at http://cor.esipfed.org/ont?iri=https://w3id.org/isample/vocabulary/specimentype/0.9/biomeaggregation might be of inetrest.
- Instantiation process.
Wd need to understand ways to map specimen <-> main define taxon (macroscopic taxon, possibly microscopic taxon)
-
Natural Product Taxonomy (with SKOS - NPClassifier)
-
NPClassifier (https://npclassifier.ucsd.edu/)
-
Classyfire (http://classyfire.wishartlab.com/)
https://www.bobdc.com/blog/skosibm/
SKOS could allow to mapp chemical taxonomies across them. Nice thing is that Classyfire classes are already mapped with CHEMONT. See http://classyfire.wishartlab.com/entities/ULGZDMOVFRHVEP-UHFFFAOYSA-N
We will get in touch with Ming (NPClassifier maintainer and DBGI computational mass spec advisor) once we have a "SKOSsed" version of NPC.
NPClassifier should be used for the taxonomy.
- MS2 spectrum and LCMS analysis
-
Laboratory object description
-
Metadata for provenance (e.g, Prov-O) https://www.w3.org/TR/prov-o/
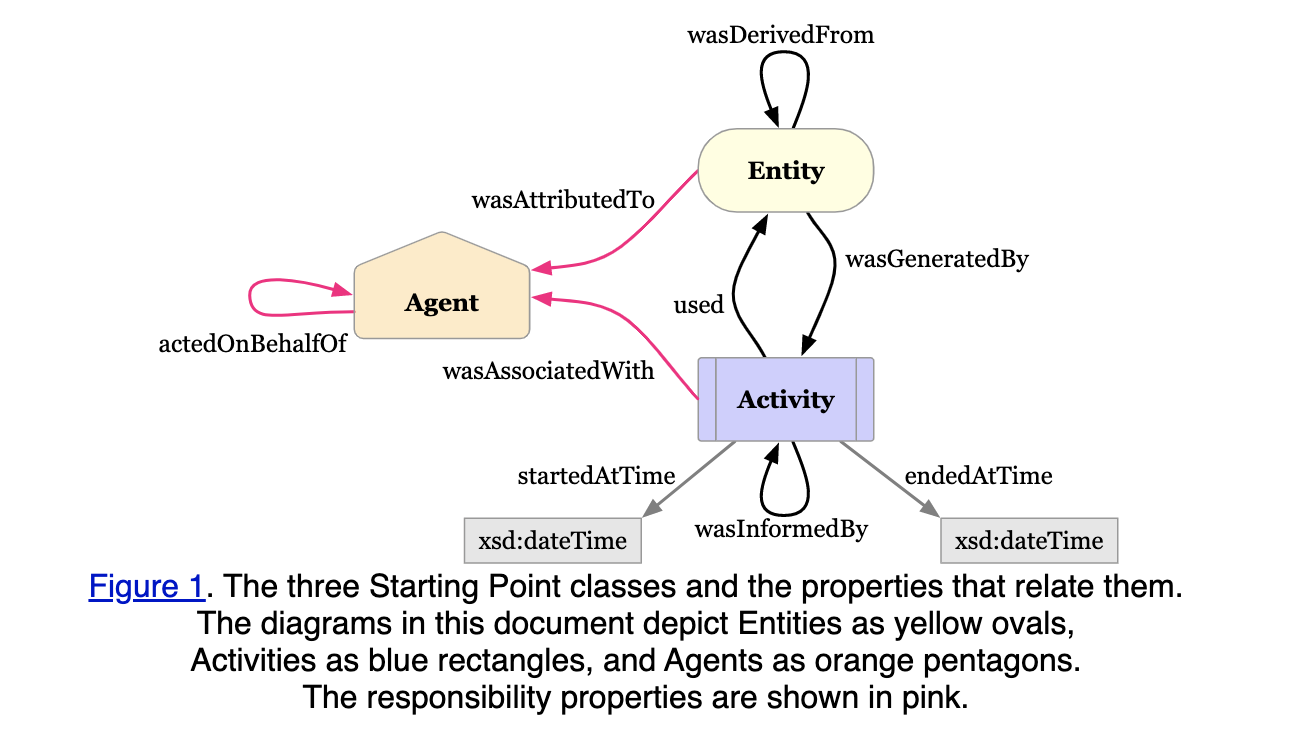
Q: could the entity be a triple ? Q: could the entity be a more complex graph ?
Luca suggests that we implement at one point quality control in the form of Continuous Integration checks.
- CI on grammatical correctness of the terms used in the model.
- CI to log the granularity of the chemical annotations / biological taxonomy completion. This would allow to understand how precisely we have defined a given spectral annotation and how precisely on organism was chracterized and this, at the full graph level.
Archived
We initially kept notes on the following pad https://mypads2.framapad.org/p/emi-semantic-model-notes-rf2evz9uw
Backlinks