methodology
Bibliography import
To import bibliography in the Dendron, you can refer to this section
Data treatment
The first step of this pilot was to clean up the list of the plants available in the Fribourg botanical garden with the help of a Python interface (for more details click here). This has permitted to obtain a clear document with all unique species with their informations and locations in the garden. In the same time we have, with the help of the "Open Tree of Life" taxonomy, created a complete taxonomy of all plants. To this base, we have linked the OTL, Wikidata and Wikipedia page for each specimen of this list. At this time, we haden't found a good protocol to assign a unique ID for each plant yet. So we have chosen to assign batch IDs that are in format DBGI_01_04_001 to DBGI_01_04_095 for the lyophilisated samples and DBGI_01_11_001 to DBGI_01_11_040 for the silica dryed ones. For the future of the project, we have decided to use different codes for the different steps of the process. For example, the sampling will be coded DBGI_SPL_XXXXXX.
Map of the Fribourg botanical garden with a drone
To have a very precise location for each harvested sample, we have made a high resolution map of the Fribourg botanical garden with a drone. This map aim to obtain centimetric coordinates precision of each sample using the Structure From Motion (SFM) technology. To see the complete description of the map creation, you can go there.
Harvesting method
The general process used for the harvesting of one sample is firstly to take a minimum of four pictures (one of the plant in general, one of the flowers/fruits/seeds/leaves in detail, one of the prelevated zone and the last of the sample with the assigned code on it) (see an example with Physalis peruviana below). These pictures are for the iNaturalist page of the sample.
 Physalis peruviana in general with it's label in the botanical garden (if available)
Physalis peruviana in general with it's label in the botanical garden (if available)
 Detailed fruits and leaves of Physalis peruviana
Detailed fruits and leaves of Physalis peruviana
 The location of the sampling indicated with the scalpel to know exactly what have been prelevated
The location of the sampling indicated with the scalpel to know exactly what have been prelevated
 Photo of the sample with the label (here Physalis peruviana, but for the next harvests, a unique code will be developed). Here the exemple has been made with the silica sampling
Photo of the sample with the label (here Physalis peruviana, but for the next harvests, a unique code will be developed). Here the exemple has been made with the silica sampling
The second step of the sampling is to add a point on the map with the open source software QGIS. You can find complete explanations and processes here. This point will be exploited later to add a more precise location of the plant as possible on iNaturalist website.
The third step of the sampling is to prelevate the part of the plant indicated on the third picture and store it in liquid nitrogen or in silica gel, according to the prefered method, that are described below.
For the sampling, we have tested two different methods, that are complementary :
- Directly put the sample in liquid Nitrogen after prelevation, and then lyophilisate in batch of approximatively 50 samples during 48 hours minimum
- Put the samples in coffee filters and then in plastic bags with silica
Liquid Nitrogen harvesting
The liquide nitrogen sample seems to be more precise, but is more difficult to apply in nature. This technic seems to be better to capt the chemicals before their degradation (oxydation for example) or disappearence (volatiles) and provide the best quality chemicals detection (devos_2007).
In fact, for the pilot, the sample is put in a folded aluminium leaf, that is directly stored in the field in liquid nitrogen. After each harvesting (approximatively 50 samples), the batch is directly put in the lyophilisator for at least 48 hours. The aluminum leafs with samples are then stored at -80°C waiting the weighting and extraction. However, for the next extractions, 15ml falcon tubes seems to be better for the harvest and storage.
Silica gel harvesting
The silica gel was first imagined for prelevating in difficult areas where liquid nitrogen is unavailable, because it's assumed to be less preservative as liquid nitrogen. But after the first tests, it seems to be as good as the lyophilization to obtain high quality samples.
In fact, for the pilot, the sample is put in a brown coffee filter, and then in a ziplock plastic bag. The silica gel is added after the harvesting session (approximatively 40 samples, that represented approximatively 5 hours in the field). Then, the silica gel is changed regularly when it becomes white. After approximatively one month, the majority of the samples are dry and don't need a silica change anymore.
Import samples on iNaturalist
After the sampling in the field, we have to import the samples on Inaturalist to identify the plant species, add the pictures and add the precise location. The advantages and utility of iNaturalist are described here
Extraction method
We have chosen to use the same extraction method for liquid nitrogen and silica gel samples to have the best comparison results.
Firstly, we prelevate 50 micrograms of dryed plants for each sample, with an error of 5%, corresponding to ±2.5 micrograms. This matter is weighted in 2ml rounded bottom eppendorf tubes, and three 4mm metal beads are added. The tubes are then stored in liquid nitrogen. The samples are given a unique code at this step, that will follow the sample during all the rest of the process. After that, the samples are passed in the Retsch machine 2.5 minutes at 25Hz. After that, 1.7ml of mix 80% methanol, 20% distillated water ans 0.1% formic acid is added, and the whole is passed in the Retsch 2.5 minutes at 25Hz again. Then, the tube is passed 2 minutes at 13'000RPM in a centrifugation machine and approximatively 1.4ml of supernatent is prelevated and stored in 1.5ml vials with non split caps. These vials are stored at -80°C.
Controls
We have made 3 different controls to see the impact of the extraction on the sampes.
- The first control has been processed exactly like the samples during the extraction phase, without plant tissues.
- The seconde control is equivalent to the first control, but has been stored 5 days at room temperature with the metal beads to see a potential impact of the rusted beads.
- The third control is just the extraction solution put on an eppedorf tube and prelevated some minutes later to put in vials for mass spectrometry analysis.
Liquid chromatography and Mass spectrometry analyse
For the passage in the liquid chromatography and the mass specrometer, 300 microliters are prelevated in the 1.5ml vials and put in an other 1.5ml vial with a 400 microliters filters with a split cap. These vials are then put in the chromatograph and analyzed by batch of maximum 270 vials.
Mass Spectrometry Data treatment
To treat the data, we have converted the raw MS data with proteowizard to then use MZmine 2.53 for the data correction. We then have imported the resluts on the [GNPS website]https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp) to create the molecular network. Finally, the molecular network has been downloaded from GNPS to display and manage it with Cytoscape 3.8.2.
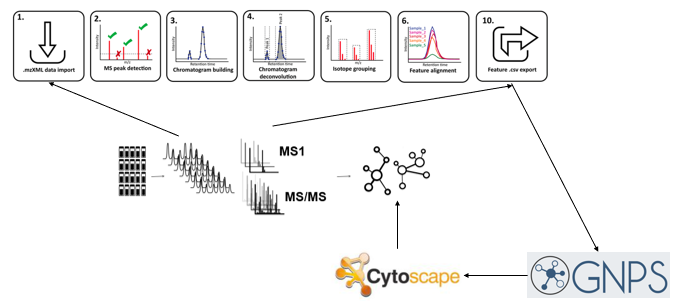
Link between the samples, the results and the database
The link between the samples, the results and the database can be resumed by the schema below.
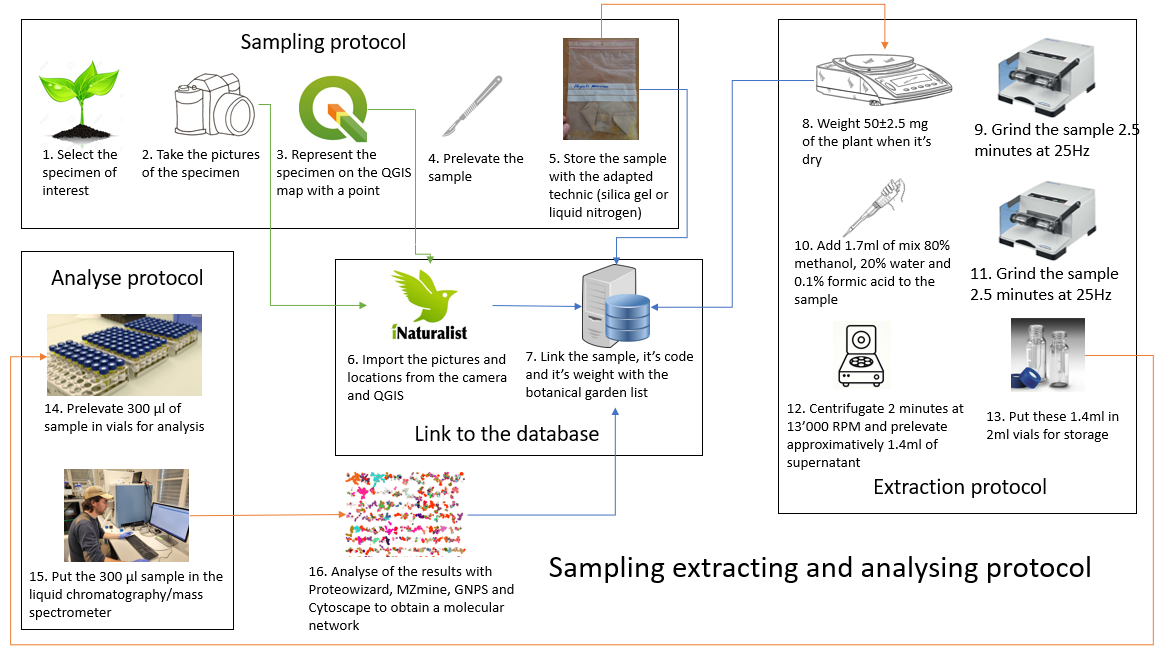 Complete schema of the pilot project
Complete schema of the pilot project
Children